The IGFA and the Future of Roosterfish
A Landmark Study

Conception

IGFA donor, traveling angler, and photographer Tom Olivo's curiosity about the roosterfish population off Costa Rica's Osa Peninsula prompted an IGFA review that revealed a significant lack of biological and ecological data on the species.
In late 2021, the IGFA was approached by Tom Olivo, an avid angler and conservationist, who sought to learn more about the local population of roosterfish in his area off the Osa Peninsula in Costa Rica. Based on those first discussions with Tom, the IGFA did a deep dive into the available biological and ecological data for roosterfish, identifying a lack of information on this recreationally important species.
After a full literature review and numerous conversations, the IGFA contacted the foremost roosterfish scientist, Dr. Sofía Ortega Garcia at CICIMAR-IPN in Mexico, who had already been working on the science needed to understand roosterfish better, in collaboration with Dr. Jaime Alvarado-Bremer of Texas A&M University. The most pressing questions that needed to be answered involved the population structure of roosterfish, whether the population was made up of one large stock or multiple subpopulations. The answer to this critical question has significant management impacts given the species had never been formally assessed or managed anywhere along its range from Baja California, Mexico, to Peru.

Dr. Sofía Ortega Garcia and Dr. Jaime Alvarado-Bremer are leading a two-year genetic study to collect fin clips from roosterfish in Mexico, Guatemala, and Costa Rica to better understand their movements and population connectivity.
Throughout our conversations with Sofía and Jaime, it was clear that genetic analysis would be the best path forward. Together with the IGFA team, Sofía and Jaime developed the ideal project to explore the level of connectivity of the population based on the genetic analysis of fin clips that could be collected through IGFA’s network of representatives, captains, lodges, and local anglers. With the project conceptualized, the IGFA could initiate a fundraising campaign to undertake the study.
Reception

To say the roosterfish fishing community embraced the study would be a gross understatement. Roosterfish anglers came out of the woodwork to assist with sample collection and help fund the study. By May of 2022, just a few short months after initiating the campaign, the research permits were in place and the two-year project officially kicked off with sample collections planned for Costa Rica, Guatemala, and Baja California, and Mexico, where some samples had already been collected in years prior by the research team. Over the next two years, over 120 fin clip samples were collected and shipped to Dr. Alvarado-Bremer’s lab in Texas for analysis. By the summer of 2024, all samples had been collected and the scientific team was putting the finishing touches on data analysis as well as compiling the results to be put into scientific manuscripts and a plain language report (see below). The IGFA is incredibly proud to have put together this project in a short time and to see results that directly impact how roosterfish should be conserved and managed.
Acknowledgments
Before we present the results of this landmark study, we want to acknowledge those who made the IGFA Roosterfish Research Project possible. We extend our deepest appreciation to The Bullard Family Foundation, courtesy of Betsy Bullard, IGFA Trustee Karen Comstock, Charles Foschini, Rocky and Judy Franich, The Guy Harvey Foundation, the International Roosterfish Tournament, Tom Olivo, PENN, Jorge Sinibaldi in Guatemala, Crocodile Bay Lodge, Bernald Pacheco from INCOPESCA in Costa Rica, and Jeffrey Feczko in Mexico for their help in making this study a success and championing this vital research. The contributions of time, expertise, and resources from the entire recreational fishing community have established a foundation for understanding and preserving these important gamefish.
The Future

Because this first project of the IGFA’s Roosterfish Research Program was a resounding success, the IGFA is currently planning a second study that will explore the foraging habits of roosterfish at different life stages to better understand the prey and habitats preferred by roosterfish as they age. Stay tuned for more information about the next project as the IGFA plans to write the book on roosterfish science with the goal of ensuring these incredible fish are available to catch for generations to come. Fundraising has already begun for this project, and we are grateful to Gary Geddes for offering to match gifts made up to $5,000 for this project. To learn more about how to support the IGFA Roosterfish Research Program, please visit the IGFA Roosterfish Research Program Website or contact IGFA Chief Development Officer Barbara E. Cini at [email protected] or (954) 498-7153.
The following report was developed by Dr. Jaime Alvarado-Bremer for the IGFA to share with the recreational fishing community. The science team, Jaime and Sofía, are currently writing a scientific manuscript to be submitted for publication.
THE POPULATION GENOMICS OF ROOSTERFISH (Nematistius pectoralis) ALONG THE EASTERN PACIFIC
Non-technical Summary
Introduction
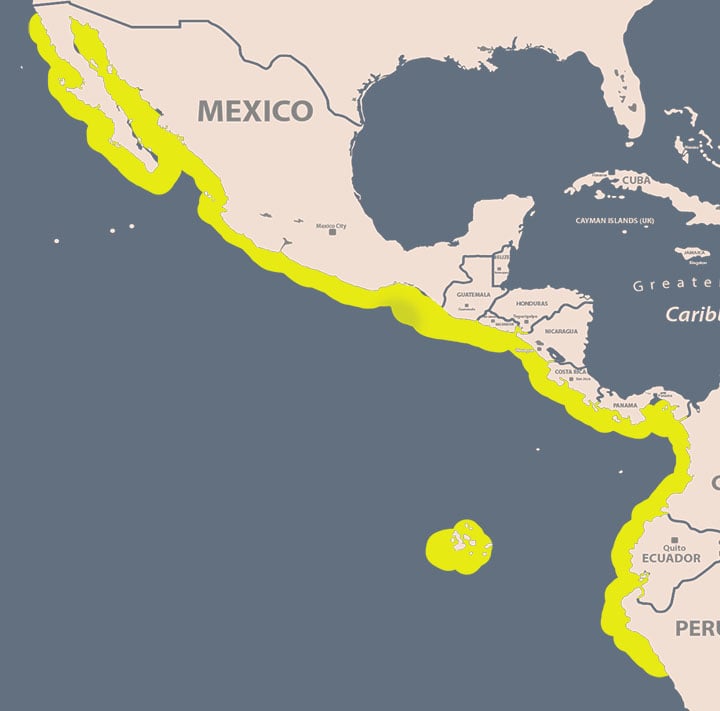
The roosterfish is highly sought after for its strong fighting abilities and striking beauty that is largely defined by the presence of a "rooster comb", formed by seven very long spines on the first dorsal fin, and by a silvery reflective and bluish-to-gray pigmentation on the head and body and four very prominent black bars, one running between their eyes, another across the posterior end of the head, with two more that begin at the base of the dorsal fin curving along the body sides towards the tail. Roosterfish are found exclusively along the warm (73-88°F; 23–31°C) shallow coastal waters of the Eastern Pacific Ocean, from San Clemente in Southern California to San Lorenzo Island, Peru, including the Gulf of California and Galapagos Island (Fig.1). They may spend most of their time between the sea surface and a depth of 12 m, and while all age classes are considered neritic, juveniles prefer the shoreline but as they mature their range includes reefs and sand bars not far from the coast.
Roosterfish are an important resource that contributes significantly to local economies favored by ecotourists such as Baja California Sur and Central America where it is targeted by inshore anglers practicing catch-and-release sportfishing, but also by artisanal local fisheries. To guarantee the future health of roosterfish populations throughout its range, it is necessary to implement conservation measures. However, the knowledge about roosterfish biology and fisheries necessary to implement sound management practices is sparse. For roosterfish, fisheries statistics are limited to catch time series, and our understanding of this species’ migrations consists of a single study. While we know that for the most part, roosterfish movements appear to be limited in scale (9-26 mi; 15 to 42 km), there is no knowledge about the level of connectivity of this species and such information is required if sound management practices are to be implemented throughout its range. Understanding the need to close this information gap, the IGFA implemented the Roosterfish Research Program (https://igfa.org/roosterfish-research-program/) focusing on answering several questions:
- What is the contemporary population structure of roosterfish throughout their range, and what are the most appropriate population management units locally and internationally?
- What is the effective population size of roosterfish?
- What is the historical gene flow of roosterfish?
- How connected is the roosterfish population? From Mexico to Peru, do roosterfish make up one large stock or multiple localized stocks?
- Should this species be managed by individual countries or as a single large stock across its range?
- What indications do we have for how roosterfish will cope with future climatic conditions along its range?
Methods
To begin answering some of these questions requires the involvement of experts in fisheries and genomics working in collaboration with sportfishermen, sport fleet operators, and government agencies assisting in obtaining the samples for genetic analysis. Here we report the successful collaboration funded by the IGFA that included Dr. Sofía Ortega García of CICIMAR, Mexico and Dr. Jaime Alvarado-Bremer of Texas A&M University (TAMU). Tissue samples for genetic analysis, which consisted of fin clips from the rooster comb (dorsal fin) as a minimally invasive approach that poses no harm to the fish, were obtained with the assistance of PISCES Sports Fleet in Mexico, Mr. Jorge Sinibaldi in Guatemala, Mr. Tom Olivo, Silena Ceballos Castro, Diego Camacho Aguilar, CONAGEBIO and INCOPESCA in Costa Rica, and numerous sportfishermen throughout the study area. As a result of this extremely successful collaborative effort, a total of 69 roosterfish samples were collected in Baja California Sur, 64 in Guatemala and 53 in Costa Rica (Fig. 2).

The genetic and genomic analyses conducted in Dr. Alvarado-Bremer’s lab at TAMU included the analysis of microsatellite markers and single nucleotide polymorphisms (SNPs). The analysis of microsatellite markers, which are short copies of DNA sequence motifs repeated in tandem, was conducted because of their strong record in identifying population structure in many species, and because a set of markers for roosterfish was available in the literature. A drawback of microsatellite analysis is the large amount of work required in the laboratory to generate size polymorphism data. Conversely, the analysis of SNPs involved generating billions of base pairs of DNA sequences for a representative sample of roosterfish from each locality, but the amount of laboratory work required is comparatively small. The major constraints in the analysis of SNPs are that it requires very high-quality DNA, compared to microsatellite markers, as well as enormous computational power to conduct bioinformatic analyses to identify SNPs. However, all these requirements are rewarded by the relative ease of obtaining thousands of SNPs several times more informative than microsatellite data.
Analyses
The analysis of nine microsatellite markers identified differences between the sample from Baja California Sur from those collected in Central America (Fig. 3). Given the large geographic distance between Baja California and Central America, which spans at least 2,200 kilometers (1367 miles), these results may not be surprising. However, identifying genetic differences in marine fishes with microsatellites is not that common because the ocean acts as a conveyor belt that facilitates contact between populations either by the active migration of fishes or due to the dispersal of eggs and larvae by ocean currents.
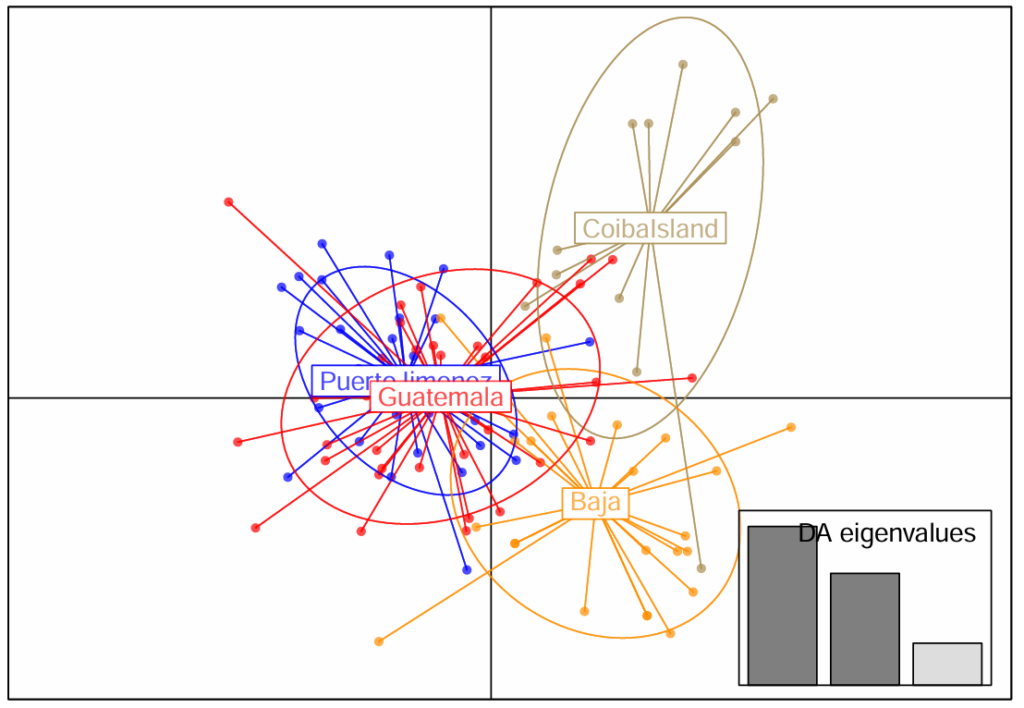
It is worth mentioning that this is the first report that identifies population structuring in roosterfish, and that result alone has scientific merit as it helps close the information gap on the biology of this species. However, there were two additional results obtained with microsatellites worth mentioning. The first, which was particularly surprising, was the genetic differentiation of Panama from the other Central American samples, which in turn appear not to be different from each other. This result is particularly puzzling given the relative proximity between Coiba Island, Panama and Puerto Jimenez, Costa Rica (200 km; 125 mi). The second surprising finding obtained with microsatellites is the apparent depauperate levels of genetic variation observed which suggest that Roosterfish populations suffered a very dramatic decline in the past that eroded large amounts of genetic variation.

This and other questions included in the IGFA’s Roosterfish Research Program will require further evaluation, some of which will be included as part of the Master of Science thesis of Ms. Emily Castillo, a graduate student conducting research at TAMU under the direction of Dr. Alvarado-Bremer, with the assistance of undergraduate student Savanna Watson (Fig. 4) who was responsible for isolating DNA and initially characterized the microsatellite markers prior to Emily’s arrival. The academic formation of future geneticists was an integral component of this project, and this was made possible with the support provided by the IGFA. More importantly, the impact on the future of these two students, largely because of their participation in this project, has been transforming. After completing her Master’s degree, Emily plans to pursue a doctorate degree in fisheries genetics, and Savanna is looking forward to starting her own Master’s of Science studies in the same field.
More importantly, the impact on the future of these two students, largely because of their participation in this project, has been transforming. After completing her Master’s degree, Emily plans to pursue a doctorate in fisheries genetics, and Savanna is looking forward to starting her own Master’s of Science studies in the same field.
The results of the analysis of SNP’s were even more informative than those obtained with microsatellites. SNP were obtained by conducting double digest Restriction Associated DNA (ddRAD) sequencing, and on average we obtained about 8.3 million reads approximately 150 bp long per individual. Accordingly, roughly 1.2 billion base pairs (bp) of high-quality DNA sequence for each of the 41 Roosterfish specimens were characterized.

The high-quality values for the sequence data are a testament to the success of the collaboration between sportfishermen and scientists. After applying multiple filters to the SNP data aimed to minimize any potential biases that would affect the interpretation of the pertinent statistics, a total of 14,366 SNPs were retained, and the results of the analysis of these data are presented in Figure 5. The most relevant finding is the strong differentiation of the samples from Baja California from the samples from Guatemala (two sampling years) and Puerto Jimenez, Costa Rica. In addition, the two Central American locals can be readily differentiated from each other, a level of differentiation not displayed by microsatellites. Further, the lack of differentiation at a temporal level for two sampling years (Y1 and Y2) in Guatemala, indicates that the genetic signals are stable across time. Unfortunately, we could not include in this analysis the sample from Panama due to the poor quality of its DNA. Future studies should assess the level of differentiation using SNPs to verify the results obtained with microsatellites.
The rejection of a single population throughout roosterfish’s range has important implications for fisheries management. The fact that each of the localities assessed was different from all others implies that the level of exchange of individuals among populations (gene flow) is extremely low. Accordingly, the data presented here suggest that each subpopulation is, for the most part, self-sustaining, and under such a scenario, if any given subpopulation were to be overexploited, it could result in population levels so low that may lead toward local extinction of Roosterfish from those waters.
Conclusions
For the first time, the existence of subpopulations of roosterfish is demonstrated. Future studies should expand to other localities throughout the roosterfish’s range to include samples from Ecuador, as well as other localities along the continental coast of Mexico (e.g., Puerto Vallarta, Ixtapa, and Puerto Escondido) and Panama. Information regarding the level of population structuring and gene flow throughout the roosterfish’s range would be particularly useful to improve the management practices of this ecologically important and charismatic fish for future generations.
Summary prepared by Dr. Jaime R. Alvarado-Bremer, Department of Marine Biology, Texas A&M University. Galveston Campus.


